TidyTuesday 2020 Week34
Plants in Danger
Preparations
library(tidyverse)
library(flair) #Highlight, Annotate, and Format your R Source CodeImport
plants <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2020/2020-08-18/plants.csv',
col_types = cols(
.default = col_double(),
binomial_name = col_character(),
country = col_character(),
continent = col_character(),
group = col_character(),
year_last_seen = col_character(),
red_list_category = col_character()
)
)
plants %>%
glimpse()## Rows: 500
## Columns: 24
## $ binomial_name <chr> "Abutilon pitcairnense", "Acaena exigua", "Acalyph…
## $ country <chr> "Pitcairn", "United States", "Congo", "Saint Helen…
## $ continent <chr> "Oceania", "North America", "Africa", "Africa", "O…
## $ group <chr> "Flowering Plant", "Flowering Plant", "Flowering P…
## $ year_last_seen <chr> "2000-2020", "1980-1999", "1940-1959", "Before 190…
## $ threat_AA <dbl> 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0,…
## $ threat_BRU <dbl> 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0,…
## $ threat_RCD <dbl> 0, 0, 0, 0, 1, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0,…
## $ threat_ISGD <dbl> 1, 1, 0, 1, 1, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ threat_EPM <dbl> 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ threat_CC <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ threat_HID <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ threat_P <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ threat_TS <dbl> 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0,…
## $ threat_NSM <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ threat_GE <dbl> 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ threat_NA <dbl> 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1,…
## $ action_LWP <dbl> 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0,…
## $ action_SM <dbl> 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ action_LP <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ action_RM <dbl> 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ action_EA <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ action_NA <dbl> 0, 0, 1, 1, 1, 0, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1,…
## $ red_list_category <chr> "Extinct in the Wild", "Extinct", "Extinct", "Exti…Tidy & Transform
case_when()
plants_by_country <- plants %>%
mutate(continent = case_when(
country == "Indonesia" ~ "Asia",
TRUE ~ continent
)
) %>%
group_by(continent, country) %>%
summarise(n = n_distinct(binomial_name), .groups = "drop")
plants_by_region <- plants_by_country %>%
mutate(
region = case_when(
country == "Cabo Verde" ~ "Cape Verde",
country == "Congo" ~ "Republic of Congo",
country == "Côte d'Ivoire" ~ "Ivory Coast",
country == "Saint Helena, Ascension and Tristan da Cunha" ~ "Ascension Island",
country == "Viet Nam" ~ "Vietnam",
country == "United Kingdom" ~ "UK",
country == "United States" ~ "USA",
country == "Pitcairn" ~ "Pitcairn Islands",
country == "Trinidad and Tobago" ~ "Trinidad",
TRUE ~ country
)
)
plants_by_region
## # A tibble: 72 x 4
## continent country n region
## <chr> <chr> <int> <chr>
## 1 Africa Angola 1 Angola
## 2 Africa Burundi 17 Burundi
## 3 Africa Cabo Verde 2 Cape Verde
## 4 Africa Cameroon 6 Cameroon
## 5 Africa Congo 6 Republic of Congo
## 6 Africa Côte d'Ivoire 2 Ivory Coast
## 7 Africa Ethiopia 3 Ethiopia
## 8 Africa Gabon 4 Gabon
## 9 Africa Guinea 14 Guinea
## 10 Africa Kenya 1 Kenya
## # … with 62 more rows
Visualise
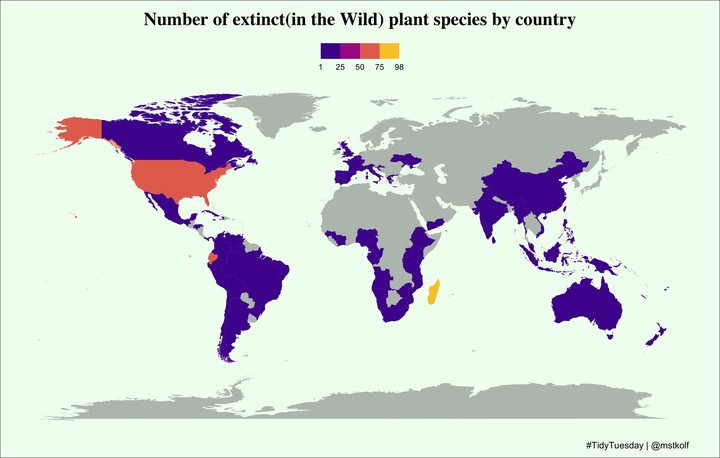
map_data("world") %>%
left_join(plants_by_region, by = "region") %>%
arrange(group, order) %>%
ggplot(aes(x = long, y = lat, group = group, fill = n, alpha = is.na(n))) +
geom_polygon() +
scale_alpha_manual(values = c(1, .5), guide = FALSE) +
scale_fill_viridis_b(
name = "", option = "plasma", breaks = c(0, 25, 50, 75, 100)
) +
theme_void() +
labs(
title = 'Number of extinct(in the Wild) plant species by country',
caption = '#TidyTuesday | @mstkolf'
) +
theme(
plot.title = element_text(face = "bold", size = 20, family = "serif", hjust = .5),
plot.background = element_rect(fill = "honeydew"),
plot.caption = element_text(size = 10, hjust = .95),
plot.margin = unit(c(.3, 0, .3, 0), "cm"),
legend.position = "top"
) +
guides(
fill = guide_colorsteps(
direction = "horizontal",
title.position = "top",
label.position = "bottom",
show.limits = TRUE
)
)
Sandbox
multiple continents
plants %>%
group_by(country) %>%
summarise(n_dist = n_distinct(continent), .groups = "drop") %>%
arrange(desc(n_dist)) %>%
top_n(10, wt = n_dist)## # A tibble: 72 x 2
## country n_dist
## <chr> <int>
## 1 Indonesia 2
## 2 Angola 1
## 3 Argentina 1
## 4 Australia 1
## 5 Belgium 1
## 6 Bermuda 1
## 7 Bhutan 1
## 8 Bolivia 1
## 9 Brazil 1
## 10 Burundi 1
## # … with 62 more rowsplants %>%
filter(country == "Indonesia") %>%
pluck("continent") %>%
unique()## [1] "Oceania" "Asia"country name
plants_by_country %>%
left_join(map_data("world"), by = c("country" = "region")) %>%
filter(is.na(long))## # A tibble: 9 x 8
## continent country n long lat group order subregion
## <chr> <chr> <int> <dbl> <dbl> <dbl> <int> <chr>
## 1 Africa Cabo Verde 2 NA NA NA NA <NA>
## 2 Africa Congo 6 NA NA NA NA <NA>
## 3 Africa Côte d'Ivoire 2 NA NA NA NA <NA>
## 4 Africa Saint Helena, Ascension … 10 NA NA NA NA <NA>
## 5 Asia Viet Nam 3 NA NA NA NA <NA>
## 6 Europe United Kingdom 1 NA NA NA NA <NA>
## 7 North Ameri… United States 66 NA NA NA NA <NA>
## 8 Oceania Pitcairn 1 NA NA NA NA <NA>
## 9 South Ameri… Trinidad and Tobago 6 NA NA NA NA <NA>extract_region <- function(long_min, long_max, lat_min, lat_max) {
map_data("world") %>%
filter(between(long, long_min, long_max) & between(lat, lat_min, lat_max)) %>%
pluck("region") %>%
unique() %>%
sort()
}
# Cabo Verde→Cape Verde
extract_region(-25, -20, 15, 18)## [1] "Cape Verde"# Congo→Republic of Congo
extract_region(10, 20, -10, -5)## [1] "Angola" "Democratic Republic of the Congo"
## [3] "Republic of Congo"# Côte d'Ivoire→Ivory Coast
extract_region(-10, 0, -10, 10)## [1] "Burkina Faso" "Ghana" "Guinea" "Ivory Coast" "Liberia"# Saint Helena, Ascension and Tristan da Cunha→Ascension Island
extract_region(-20, -10, -50, -5)## [1] "Ascension Island"# Viet Nam→Vietnam
extract_region(105, 110, 10, 20)## [1] "Cambodia" "China" "Laos" "Thailand" "Vietnam"# United Kingdom→UK
extract_region(-5, 5, 50, 55)## [1] "Belgium" "France" "Isle of Man" "Netherlands" "UK"# United States→USA
extract_region(-110, -90, 25, 30)## [1] "Mexico" "USA"# Pitcairn→Pitcairn Islands
extract_region(-130, -120, -30, -20)## [1] "Pitcairn Islands"# Trinidad and Tobago→Trinidad
extract_region(-65, -60, 11, 13)## [1] "Grenada" "Grenadines" "Tobago" "Venezuela"plants_by_region %>%
left_join(map_data("world"), by = "region") %>%
filter(is.na(long))## # A tibble: 0 x 9
## # … with 9 variables: continent <chr>, country <chr>, n <int>, region <chr>,
## # long <dbl>, lat <dbl>, group <dbl>, order <int>, subregion <chr>References
Bodwin, Kelly, and Hunter Glanz. 2021. Flair: Highlight, Annotate, and Format Your r Source Code.
Wickham, Hadley. 2019. Tidyverse: Easily Install and Load the ’Tidyverse’. https://CRAN.R-project.org/package=tidyverse.
Reproducibility
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.0.3 (2020-10-10)
## os macOS Big Sur 10.16
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate ja_JP.UTF-8
## ctype ja_JP.UTF-8
## tz Asia/Tokyo
## date 2021-05-03
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date lib source
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.0)
## backports 1.1.7 2020-05-13 [1] CRAN (R 4.0.0)
## blogdown 1.3 2021-04-14 [1] CRAN (R 4.0.2)
## bookdown 0.20 2020-06-23 [1] CRAN (R 4.0.2)
## broom 0.7.4 2021-01-29 [1] CRAN (R 4.0.2)
## cellranger 1.1.0 2016-07-27 [1] CRAN (R 4.0.0)
## cli 2.2.0 2020-11-20 [1] CRAN (R 4.0.2)
## colorspace 1.4-1 2019-03-18 [1] CRAN (R 4.0.0)
## crayon 1.4.0 2021-01-30 [1] CRAN (R 4.0.3)
## curl 4.3 2019-12-02 [1] CRAN (R 4.0.0)
## DBI 1.1.0 2019-12-15 [1] CRAN (R 4.0.0)
## dbplyr 2.1.0 2021-02-03 [1] CRAN (R 4.0.3)
## digest 0.6.27 2020-10-24 [1] CRAN (R 4.0.2)
## dplyr * 1.0.4 2021-02-02 [1] CRAN (R 4.0.2)
## ellipsis 0.3.1 2020-05-15 [1] CRAN (R 4.0.0)
## evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.0)
## fansi 0.4.2 2021-01-15 [1] CRAN (R 4.0.2)
## farver 2.0.3 2020-01-16 [1] CRAN (R 4.0.0)
## flair * 0.0.2 2021-02-23 [1] Github (kbodwin/flair@1232fd5)
## forcats * 0.5.1 2021-01-27 [1] CRAN (R 4.0.2)
## fs 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
## generics 0.1.0 2020-10-31 [1] CRAN (R 4.0.2)
## ggplot2 * 3.3.3 2020-12-30 [1] CRAN (R 4.0.2)
## glue 1.4.2 2020-08-27 [1] CRAN (R 4.0.2)
## gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.0)
## haven 2.3.1 2020-06-01 [1] CRAN (R 4.0.2)
## here 1.0.1 2020-12-13 [1] CRAN (R 4.0.2)
## hms 1.0.0 2021-01-13 [1] CRAN (R 4.0.2)
## htmltools 0.5.0 2020-06-16 [1] CRAN (R 4.0.2)
## httr 1.4.2 2020-07-20 [1] CRAN (R 4.0.2)
## jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.0.2)
## knitr 1.28 2020-02-06 [1] CRAN (R 4.0.0)
## labeling 0.3 2014-08-23 [1] CRAN (R 4.0.0)
## lifecycle 1.0.0 2021-02-15 [1] CRAN (R 4.0.2)
## lubridate 1.7.9.2 2020-11-13 [1] CRAN (R 4.0.2)
## magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.0.2)
## maps 3.3.0 2018-04-03 [1] CRAN (R 4.0.2)
## modelr 0.1.8 2020-05-19 [1] CRAN (R 4.0.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.0)
## pillar 1.4.7 2020-11-20 [1] CRAN (R 4.0.2)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.0)
## purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.0.0)
## R6 2.5.0 2020-10-28 [1] CRAN (R 4.0.2)
## Rcpp 1.0.4.6 2020-04-09 [1] CRAN (R 4.0.0)
## readr * 1.4.0 2020-10-05 [1] CRAN (R 4.0.2)
## readxl 1.3.1 2019-03-13 [1] CRAN (R 4.0.0)
## reprex 1.0.0 2021-01-27 [1] CRAN (R 4.0.2)
## rlang 0.4.10 2020-12-30 [1] CRAN (R 4.0.2)
## rmarkdown 2.6 2020-12-14 [1] CRAN (R 4.0.2)
## rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.0.2)
## rstudioapi 0.11 2020-02-07 [1] CRAN (R 4.0.0)
## rvest 1.0.0 2021-03-09 [1] CRAN (R 4.0.2)
## scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.0)
## sessioninfo * 1.1.1 2018-11-05 [1] CRAN (R 4.0.2)
## stringi 1.4.6 2020-02-17 [1] CRAN (R 4.0.0)
## stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.0.0)
## tibble * 3.0.6 2021-01-29 [1] CRAN (R 4.0.2)
## tidyr * 1.1.2 2020-08-27 [1] CRAN (R 4.0.2)
## tidyselect 1.1.0 2020-05-11 [1] CRAN (R 4.0.0)
## tidyverse * 1.3.0 2019-11-21 [1] CRAN (R 4.0.0)
## utf8 1.1.4 2018-05-24 [1] CRAN (R 4.0.0)
## vctrs 0.3.6 2020-12-17 [1] CRAN (R 4.0.2)
## viridisLite 0.3.0 2018-02-01 [1] CRAN (R 4.0.0)
## withr 2.4.1 2021-01-26 [1] CRAN (R 4.0.2)
## xfun 0.22 2021-03-11 [1] CRAN (R 4.0.2)
## xml2 1.3.2 2020-04-23 [1] CRAN (R 4.0.0)
## yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.0/Resources/library